Abstract¶
The Control Module of the Developer Cell focuses on achieving temporal regulation of protein production using the PURE system. While the PURE cytosol lacks proteases—offering an advantage in some contexts—it also presents a limitation for controlling system dynamics. For instance, when a sensor in the PURE cytosol is activated, it may remain permanently on since the output reporter protein is not degraded. As synthetic cell researchers advance toward more complex modules and cells, the ability to regulate protein degradation becomes essential for functions like level matching, homeostasis, and time-dependent behaviors, such as cell cycle regulation. To address this, we will integrate the ClpXP protease, enabling the continuous degradation of targeted proteins.
Overview¶
The Control Module of the Developer Cell is dedicated to enabling precise, time-resolved control of protein expression within synthetic minimal systems, particularly those based on the PURE system. The PURE system offers a defined, protease-free biochemical environment that allows high-fidelity transcription and translation without the background interference found in crude cell lysates. While this protease-free context is beneficial for maximizing protein yield and reducing unwanted degradation, it introduces a critical limitation: the lack of mechanisms for post-translational regulation, particularly controlled protein degradation.
This limitation becomes increasingly significant as synthetic cell engineering moves toward more dynamic and autonomous behaviors. Without an active degradation pathway, any protein expressed in response to a stimulus such as a sensor activation remains indefinitely active, even after the stimulus is removed. This impedes the design of synthetic circuits requiring time-dependent or reversible responses, including feedback regulation, homeostasis, noise filtering, signal reset, and periodic behaviors akin to cell cycle progression (McGinness 2006).
To overcome this challenge, the Control Module aims to incorporate a programmable degradation pathway using the ClpXP protease system (Figure 1), allowing researchers to actively manage protein lifetimes in the synthetic environment. By integrating ClpXP into the PURE-based Developer Cell, we build on the work of Niederholtmeyer et al. and introduce the capacity for selective, energy-dependent degradation of target proteins, enabling dynamic control over circuit behavior and opening the door to more sophisticated synthetic cellular functionalities.
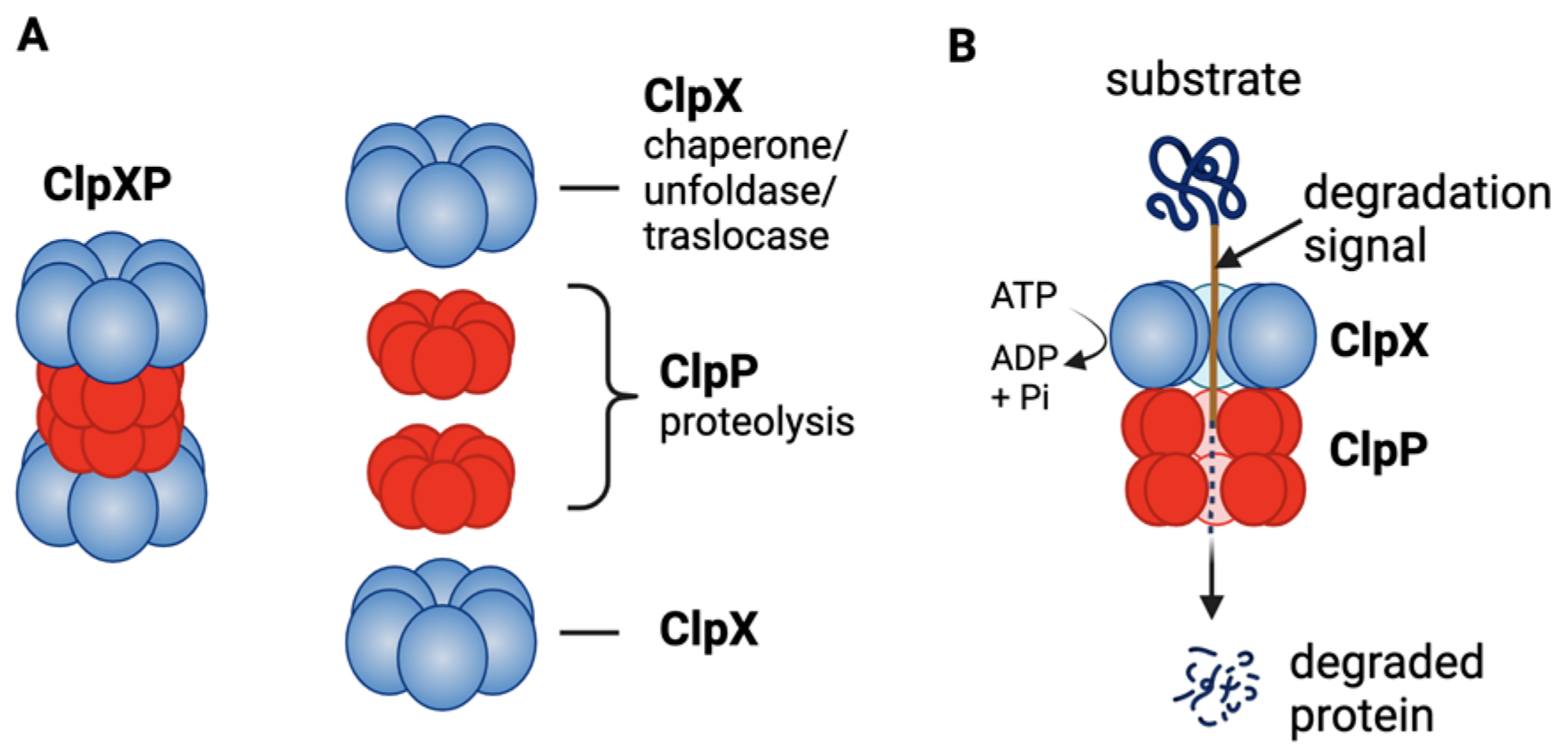
Figure 1:ClpXP structure and function: A) ClpXP consists of ClpX (ATPase) and ClpP; B) ClpX recognizes degrons of target protein, unfolds the proteins and threads them into ClpP for degradation. Figure by Wedam et al. used under CC-BY-4.0 / cropped from original.
Background¶
ClpXP is a highly conserved, ATP-dependent proteolytic complex that plays a central role in protein quality control and regulatory degradation in prokaryotic cells. It is composed of two main components: the ClpX ATPase, which recognizes and unfolds substrate proteins, and the ClpP peptidase, which degrades the unfolded polypeptides into short peptides (Baker 2011). This two-part system is capable of tightly regulated and substrate-specific degradation, a feature that is critical for synthetic systems requiring fine-tuned control over protein turnover.
In the context of the PURE system, ClpXP has been functionally reconstituted and shown to maintain activity when supplied with appropriate ATP levels Niederholtmeyer et al. (2013). Importantly, ClpXP exhibits high specificity by recognizing substrate proteins that carry a C-terminal ssrA degradation tag, allowing researchers to selectively mark proteins for degradation while leaving other components of the system untouched. This specificity ensures that only user-defined targets are removed from the system, avoiding unwanted disruption of essential cellular processes encoded within the synthetic cytosol.
Furthermore, ClpXP’s reliance on ATP hydrolysis couples its activity to the metabolic state of the system. This makes it an ideal candidate for integration with the energy module of the Developer Cell platform, ensuring coordinated control over both protein synthesis and degradation. Together, these features make ClpXP a powerful tool for implementing regulated, dynamic behavior in synthetic cells, bringing us closer to building minimal cells with lifelike properties such as adaptability, robustness, and controlled developmental trajectories.
Approach and success criteria¶
The approach we will take to validating the successful implementation of the ClpXP module within the Developer Cell will involve the use of both purified proteins and cell-free expressed proteins. This will introduce a number of OpenMTA plasmids to the Nucleus Distribution (see Table 1). The work will generally proceed from validating components in bulk reactions before testing in synthetic cells. The key objectives and tasks that will define the success of this work are as follows:
- Validate cell-free expression of ClpXP: Use the PURE system to express
pOpen-pT7-ClpP-CHisandpOpen-pT7-ClpX-CHis, then assess whether the purifieddeGFP-CHis-ssrAcan be degraded by monitoring the decrease in green fluorescence over time. - Validate purified ClpXP: Use the PURE system to express
pOpen-pT7-deGFP-ssrA, then add purified ClpX and ClpP proteins to determine whether green fluorescence decreases over time, indicating degradation. - Control dynamics of PURExpression: Co-express
pOpen-pT7-ClpP-CHis,pOpen-pT7-ClpX-CHis, andpOpen-pT7-deGFP-ssrAin the PURE system, and tune the DNA concentrations to determine whether GFP fluorescence can be: 1) Maintained at a steady level over time and 2) reduced over time.
Table 1:List of plasmid constructs that will be used to evaluate the ClpXP Control Module.
| For cell-free expression | For protein purification |
|---|---|
pOpen-pT7-ClpP-CHis | pET28a-ClpP-CHis |
pOpen-pT7-ClpX-linker-CHis | pET28a-ClpX-linker-CHis |
pOpen-pT7-ClpX-CHis | pET28a-ClpX-CHis |
pOpen-pT7-deGFP-CHis-ssrA | pET28a-deGFP-CHis-ssrA |
pOpen-pT7-deGFP-ssrA | pET28a-deGFP-ssrA |
pOpen-pT7-plamGFP-CHis-ssrA | pET28a-plamGFP-CHis-ssrA |
pOpen-pT7-sspB-CHis | pET28a-sspB-CHis |
Expected challenges and new horizons¶
We anticipate some challenges with developing the purified ClpXP proteins. For example, ClpX and ClpP are endogenous proteins and may represent a challenge for overexpression in E. coli. Additionally, deGFP-ssrA may be difficult to express because it is susceptible to degradation by the endogenous ClpXP system. Meanwhile, simultaneously tuning three independent DNA templates in a single PURE reaction will add another level of complexity. Accomplishing the goals described above will enable us to proceed with integrating the ClpXP-based Control Module with the other Developer Cell Modules.
Acknowledgments
This work is supported by the Astera Institute and Sloan Foundation (Grant G-2024-22735).
- McGinness, K. E., Baker, T. A., & Sauer, R. T. (2006). Engineering Controllable Protein Degradation. Molecular Cell, 22(5), 701–707. 10.1016/j.molcel.2006.04.027
- Niederholtmeyer, H., Stepanova, V., & Maerkl, S. J. (2013). Implementation of cell-free biological networks at steady state. Proceedings of the National Academy of Sciences, 110(40), 15985–15990. 10.1073/pnas.1311166110
- Wedam, R., Greer, Y. E., Wisniewski, D. J., Weltz, S., Kundu, M., Voeller, D., & Lipkowitz, S. (2023). Targeting Mitochondria with ClpP Agonists as a Novel Therapeutic Opportunity in Breast Cancer. Cancers, 15(7), 1936. 10.3390/cancers15071936
- Baker, T. A., & Sauer, R. T. (2012). ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1823(1), 15–28. 10.1016/j.bbamcr.2011.06.007