MTHFS - an Unexpected Enzyme in PURE
Why Folinic Acid Needs Extra Help
Contents
Analysis (20250220)
!pip install nucleus-cdk | tail -n2%load_ext autoreload
%autoreload 2
from cdk.analysis.cytosol import platereader as pr
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
pr.plot_setup()data, platemap = pr.load_platereader_data(
data_file="../02-data/20250220-MTHFS-02.txt",
platemap_file="../01-design/platemap.tsv",
platereader="biotek"
)
data Well Row Column Time Seconds Temperature (C) \
0 D2 D 2 0 days 00:00:00 0.00 37.00
1 D2 D 2 0 days 00:05:00 300.00 37.00
2 D2 D 2 0 days 00:10:00 600.00 37.00
3 D2 D 2 0 days 00:15:00 900.00 37.00
4 D2 D 2 0 days 00:20:00 1200.00 37.00
... ... .. ... ... ... ...
1085 D20 D 20 0 days 08:40:00 31200.00 37.00
1086 D20 D 20 0 days 08:45:00 31500.00 37.00
1087 D20 D 20 0 days 08:50:00 31800.00 37.00
1088 D20 D 20 0 days 08:55:00 32100.00 37.00
1089 D20 D 20 0 days 09:00:00 32400.00 37.00
Read Data Sample # Name ... SMS-03-06 \
0 485/20,528/20 0.00 1 no folinic acid ... 4.00
1 485/20,528/20 7.00 1 no folinic acid ... 4.00
2 485/20,528/20 17.00 1 no folinic acid ... 4.00
3 485/20,528/20 30.00 1 no folinic acid ... 4.00
4 485/20,528/20 40.00 1 no folinic acid ... 4.00
... ... ... ... ... ... ...
1085 485/20,528/20 5.00 10 Negative ... NaN
1086 485/20,528/20 5.00 10 Negative ... NaN
1087 485/20,528/20 6.00 10 Negative ... NaN
1088 485/20,528/20 6.00 10 Negative ... NaN
1089 485/20,528/20 6.00 10 Negative ... NaN
bnext p-mix-03-03 NEB Solution A NEB solution B RNAse Inhibitor \
0 1.30 NaN NaN 0.40
1 1.30 NaN NaN 0.40
2 1.30 NaN NaN 0.40
3 1.30 NaN NaN 0.40
4 1.30 NaN NaN 0.40
... ... ... ... ...
1085 NaN 4.00 3.00 0.00
1086 NaN 4.00 3.00 0.00
1087 NaN 4.00 3.00 0.00
1088 NaN 4.00 3.00 0.00
1089 NaN 4.00 3.00 0.00
DNA (deGFP) H2O MTHFS 0.2 mM 5,10-methenyl-THF Total Volume
0 0.50 1.00 0.00 NaN 10.00
1 0.50 1.00 0.00 NaN 10.00
2 0.50 1.00 0.00 NaN 10.00
3 0.50 1.00 0.00 NaN 10.00
4 0.50 1.00 0.00 NaN 10.00
... ... ... ... ... ...
1085 0.00 3.00 NaN NaN 10.00
1086 0.00 3.00 NaN NaN 10.00
1087 0.00 3.00 NaN NaN 10.00
1088 0.00 3.00 NaN NaN 10.00
1089 0.00 3.00 NaN NaN 10.00
[1090 rows x 23 columns]pr.plot_plate(data)<seaborn.axisgrid.FacetGrid at 0x6ffcdf959d60>
names_to_remove = ['5,10-methenyl-THF',
'folinic + BE MTHFS (1 uL)',
'folinic + BE MTHFS (1.61 uL)',
'NEB ribosome control']
replace_dict = {'no folinic acid':'no Folinic Acid',
'no folinic + 5,10-methenyl-THF':'5,10-methenyl-THF',
'folinic':'Folinic Acid',
'folinic + MTHFS (0.5 uL)':'Folinic Acid + MTHFS'}
custom_order = ['no Folinic Acid',
'Folinic Acid',
'Folinic Acid + MTHFS',
'5,10-methenyl-THF',
'Positive',
'Negative']
data_drop = data.drop(data[data['Name'].isin(names_to_remove)].index)
data_drop['Name'] = data_drop['Name'].replace(replace_dict)# Generate the plot (assuming `pr.plot_curves` returns a FacetGrid object)
sns.set_palette('colorblind')
g = pr.plot_curves(data_drop, hue_order=custom_order)
g.set_xticklabels(rotation=30)
plt.xlim([0,0.35])
plt.savefig("MTHFS_kinetics.png");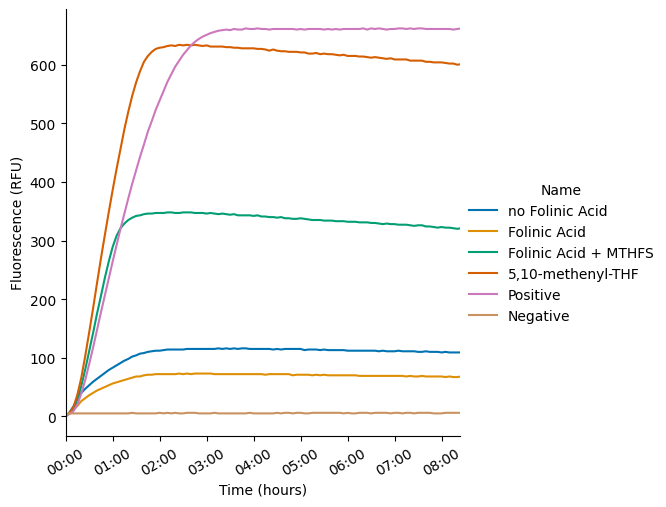
pr.plot_steadystate(data_drop, order=custom_order, palette='colorblind')
plt.savefig("MTHFS_endpoint.png");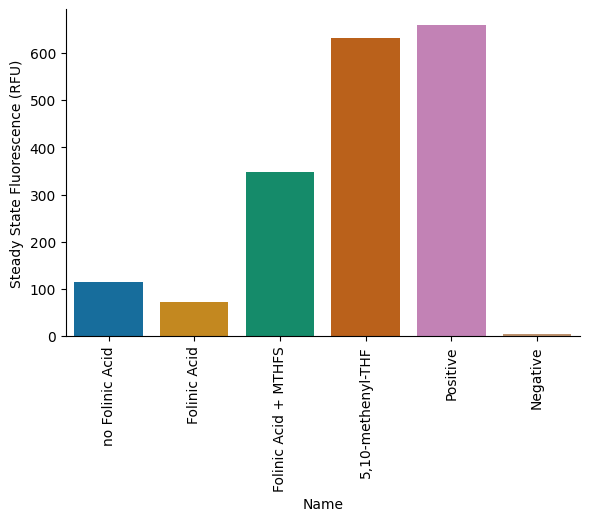
kinetics = pr.kinetic_analysis(data_drop)kinetics = pr.kinetic_analysis(data)
pr.plot_kinetics(data, kinetics)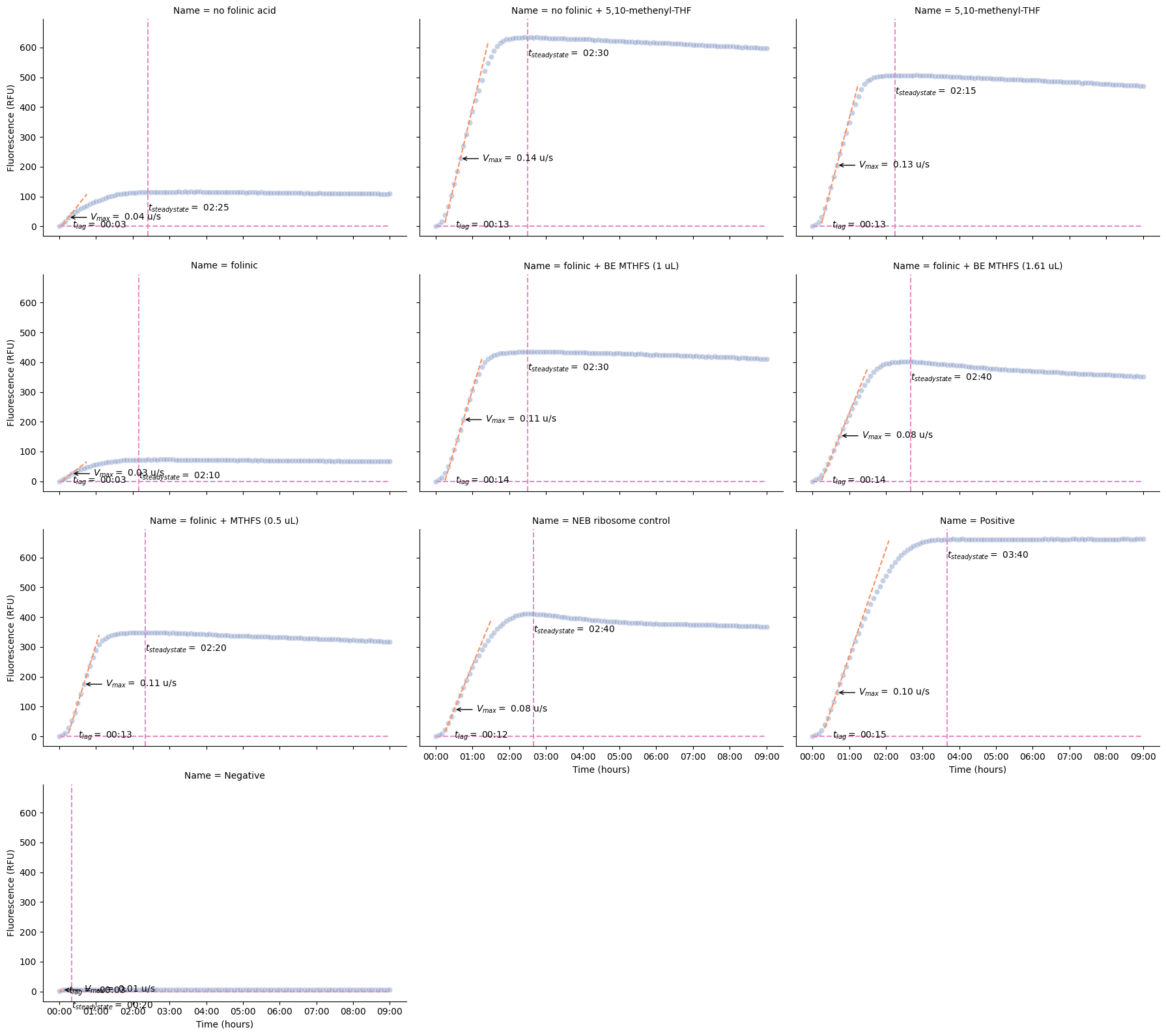
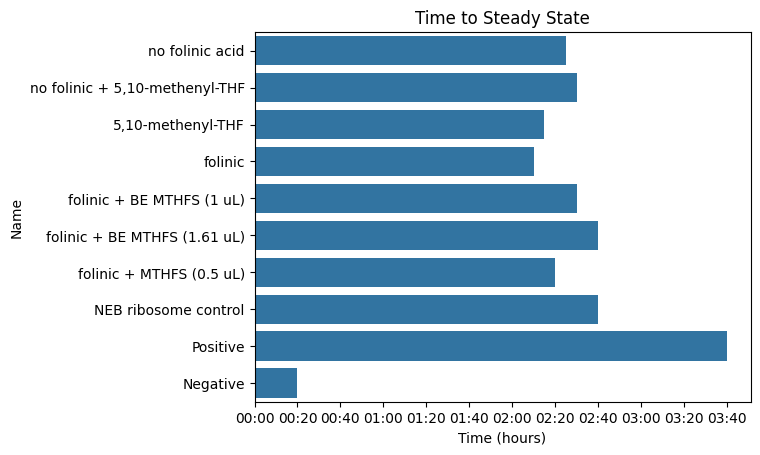
ax = sns.barplot(data=kinetics["Steady State"], y="Name", x="Time", orient="y")
ax.set_title("Time to Steady State")
pr._plot_timedelta(ax)
ax = sns.barplot(data=kinetics["Velocity"], y="Name", x="Max", orient="y")
ax.set_title("Maximum velocity")
ax.set_xlabel("$V_{max}$ (u/s)")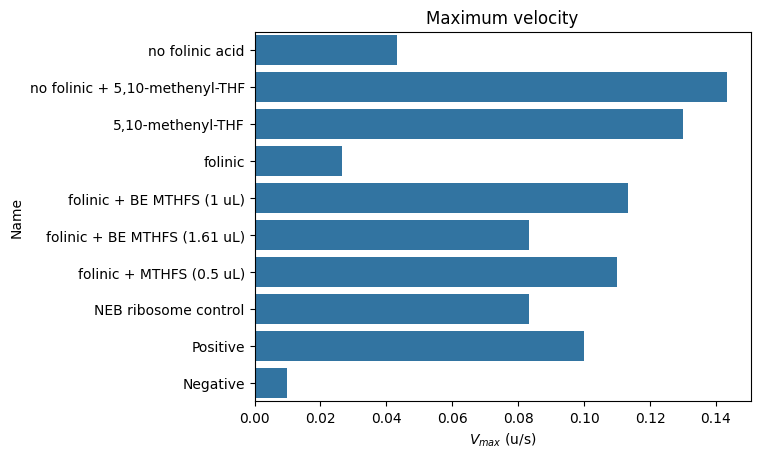
Annotate plots¶
Format time¶
import pandas as pd
def format_timedelta(td):
if pd.isna(td):
return ''
# Convert to seconds
total_seconds = td.total_seconds()
# Calculate hours, minutes, seconds
hours = int(total_seconds // 3600)
minutes = int((total_seconds % 3600) // 60)
seconds = int(total_seconds % 60)
# Format as HH:MM:SS
return f"{hours:02d}:{minutes:02d}:{seconds:02d}"
bonk = kinetics.xs("Time", level=1, axis=1).map(format_timedelta)
bonk.to_excel("bonk-time.xlsx")kinetics Velocity \
Time Data Max
Well Name Read
D2 no folinic acid 485/20,528/20 0 days 00:15:00 30.00 0.04
D4 no folinic + 5,10-methenyl-THF 485/20,528/20 0 days 00:40:00 227.00 0.14
D6 5,10-methenyl-THF 485/20,528/20 0 days 00:40:00 205.00 0.13
D8 folinic 485/20,528/20 0 days 00:20:00 26.00 0.03
D10 folinic + BE MTHFS (1 uL) 485/20,528/20 0 days 00:45:00 207.00 0.11
D12 folinic + BE MTHFS (1.61 uL) 485/20,528/20 0 days 00:45:00 153.00 0.08
D14 folinic + MTHFS (0.5 uL) 485/20,528/20 0 days 00:40:00 175.00 0.11
D16 NEB ribosome control 485/20,528/20 0 days 00:30:00 90.00 0.08
D18 Positive 485/20,528/20 0 days 00:40:00 147.00 0.10
D20 Negative 485/20,528/20 0 days 00:05:00 5.00 0.01
Lag \
Time
Well Name Read
D2 no folinic acid 485/20,528/20 0 days 00:03:27.692307692
D4 no folinic + 5,10-methenyl-THF 485/20,528/20 0 days 00:13:36.279069767
D6 5,10-methenyl-THF 485/20,528/20 0 days 00:13:43.076923077
D8 folinic 485/20,528/20 0 days 00:03:45
D10 folinic + BE MTHFS (1 uL) 485/20,528/20 0 days 00:14:33.529411765
D12 folinic + BE MTHFS (1.61 uL) 485/20,528/20 0 days 00:14:24
D14 folinic + MTHFS (0.5 uL) 485/20,528/20 0 days 00:13:29.090909091
D16 NEB ribosome control 485/20,528/20 0 days 00:12:00
D18 Positive 485/20,528/20 0 days 00:15:30
D20 Negative 485/20,528/20 -1 days +23:56:40
Steady State \
Data Time Data
Well Name Read
D2 no folinic acid 485/20,528/20 0.00 0 days 02:25:00 114.00
D4 no folinic + 5,10-methenyl-THF 485/20,528/20 0.00 0 days 02:30:00 633.00
D6 5,10-methenyl-THF 485/20,528/20 0.00 0 days 02:15:00 506.00
D8 folinic 485/20,528/20 0.00 0 days 02:10:00 72.00
D10 folinic + BE MTHFS (1 uL) 485/20,528/20 0.00 0 days 02:30:00 434.00
D12 folinic + BE MTHFS (1.61 uL) 485/20,528/20 0.00 0 days 02:40:00 401.00
D14 folinic + MTHFS (0.5 uL) 485/20,528/20 0.00 0 days 02:20:00 347.00
D16 NEB ribosome control 485/20,528/20 0.00 0 days 02:40:00 411.00
D18 Positive 485/20,528/20 0.00 0 days 03:40:00 660.00
D20 Negative 485/20,528/20 0.00 0 days 00:20:00 5.00
Fit
L k x0
Well Name Read
D2 no folinic acid 485/20,528/20 0 0 0
D4 no folinic + 5,10-methenyl-THF 485/20,528/20 0 0 0
D6 5,10-methenyl-THF 485/20,528/20 0 0 0
D8 folinic 485/20,528/20 0 0 0
D10 folinic + BE MTHFS (1 uL) 485/20,528/20 0 0 0
D12 folinic + BE MTHFS (1.61 uL) 485/20,528/20 0 0 0
D14 folinic + MTHFS (0.5 uL) 485/20,528/20 0 0 0
D16 NEB ribosome control 485/20,528/20 0 0 0
D18 Positive 485/20,528/20 0 0 0
D20 Negative 485/20,528/20 0 0 0